Experimental approaches
In vivo Ca2+ imaging
The lab mainly use miniscope for Ca2+ imaging experiments. The miniscope is a compact, head-mounted fluorescence microscope designed for in vivo Ca²⁺ imaging in freely moving mice. It enables real-time observation of neuronal activity at the cellular level, offering high spatial and temporal resolution. The miniscope’s lightweight and portable design minimizes behavioral interference, making it ideal for studying neural circuits in naturalistic settings. It allows for long-term recordings in freely behaving animals, capturing the dynamics of neural activity during complex tasks.
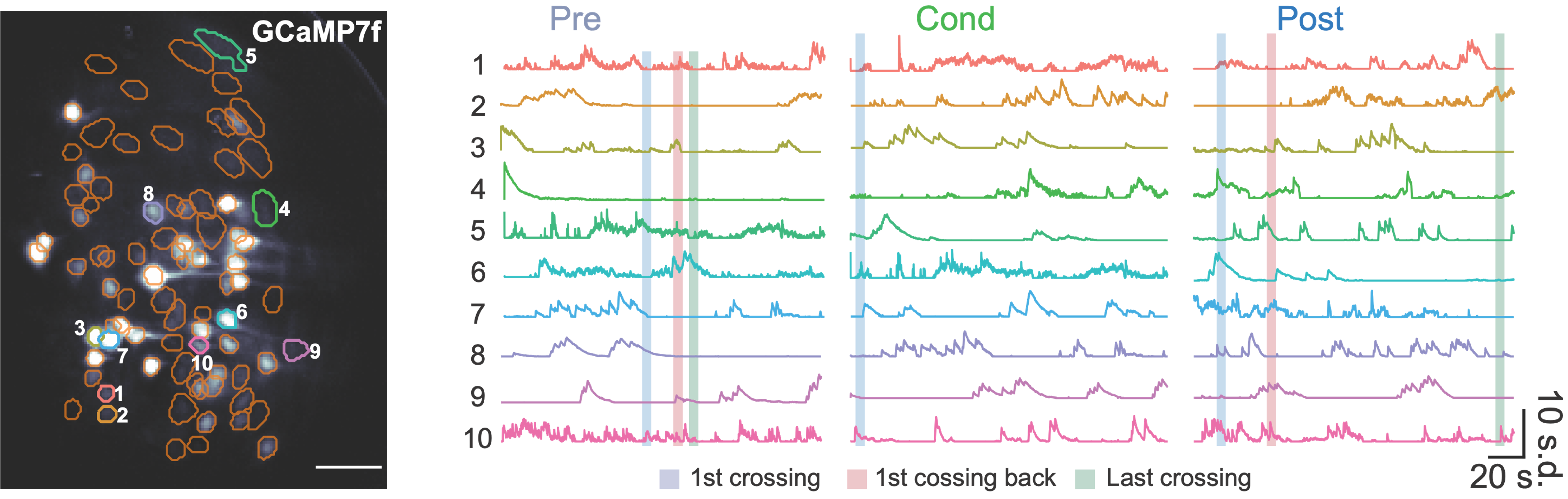 |
Electrophysiology (paired recording)
Whole-cell patch clamp is a technique to study ion channel activity and cellular electrical properties. Paired whole-cell recordings, where two cells are simultaneously patched, allow precise analysis of synaptic transmission and connectivity. This approach enables researchers to measure pre- and postsynaptic responses in real-time, offering insights into circuit dynamics, synaptic strength, and plasticity. Advantages include high temporal and spatial resolution, the ability to control intracellular environments, and direct access to synaptic interactions. It is particularly valuable in neuroscience for investigating synaptic transmission, synaptic plasticity, and functional connectivity.
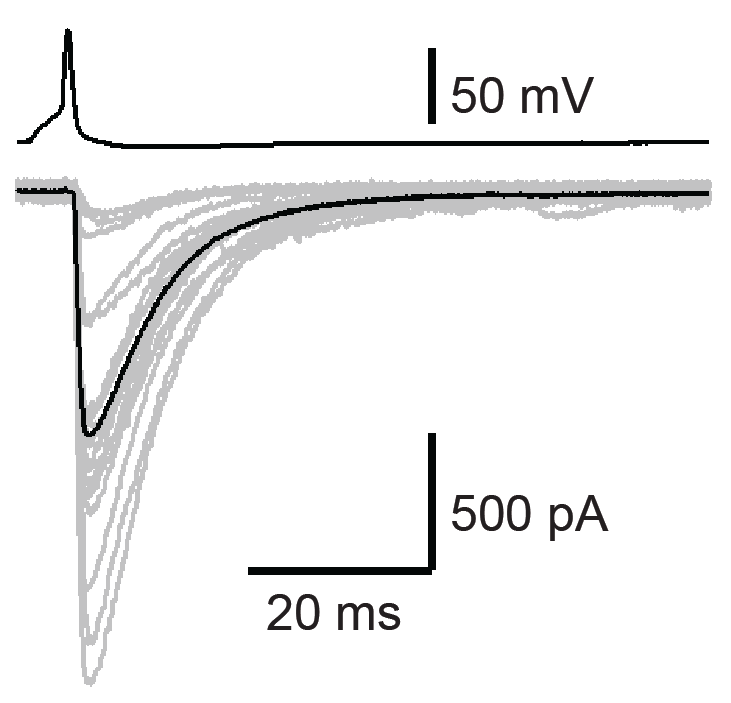
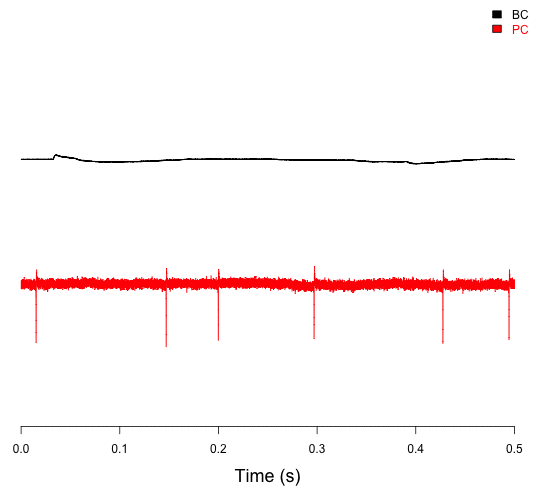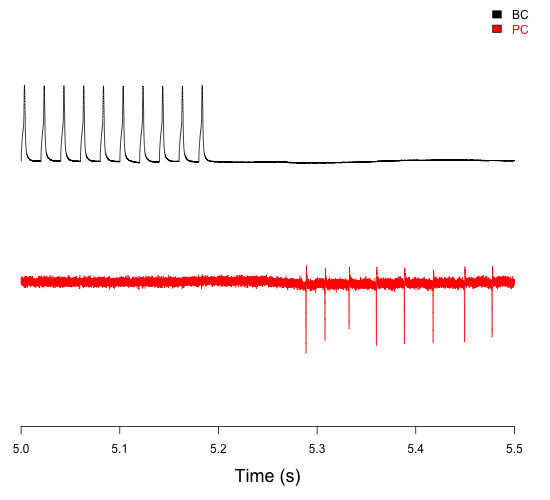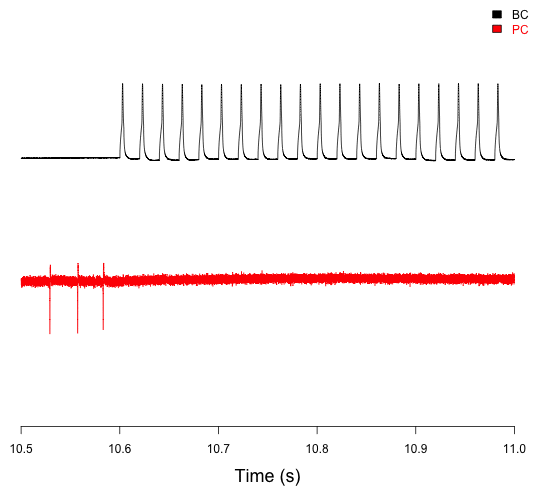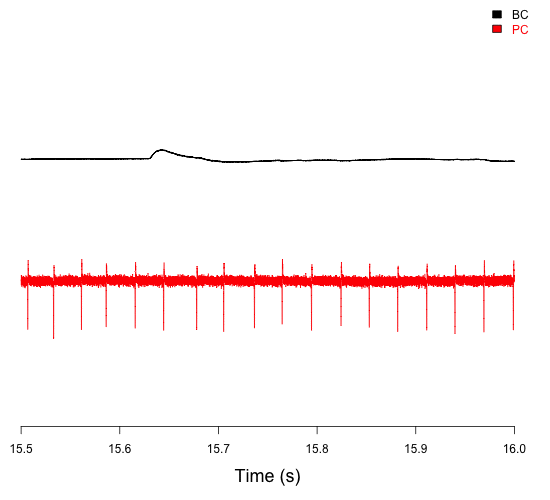
Paired recording of connected basket cells (BC) and Purkinje cells (PC) in the cerebellum, showing both their activity and how BC firing influences PC firing (Chen et al., 2017a,b).
Viral neuronal tracing
Viral tracing is a powerful technique to map neural circuits by utilizing genetically engineered viruses to label neurons and their connections. Anterograde tracing labels downstream targets by transporting the virus from the cell body along axons to synaptic terminals, revealing outputs of specific neurons. Retrograde tracing, on the other hand, maps inputs by transporting the virus from axon terminals back to the originating cell bodies, identifying presynaptic neurons. Advanced viral tools, such as rabies and AAV vectors, enable cell-type-specific tracing, functional manipulation, and circuit mapping at single-neuron resolution, providing insights into the organization and connectivity of complex neural networks.
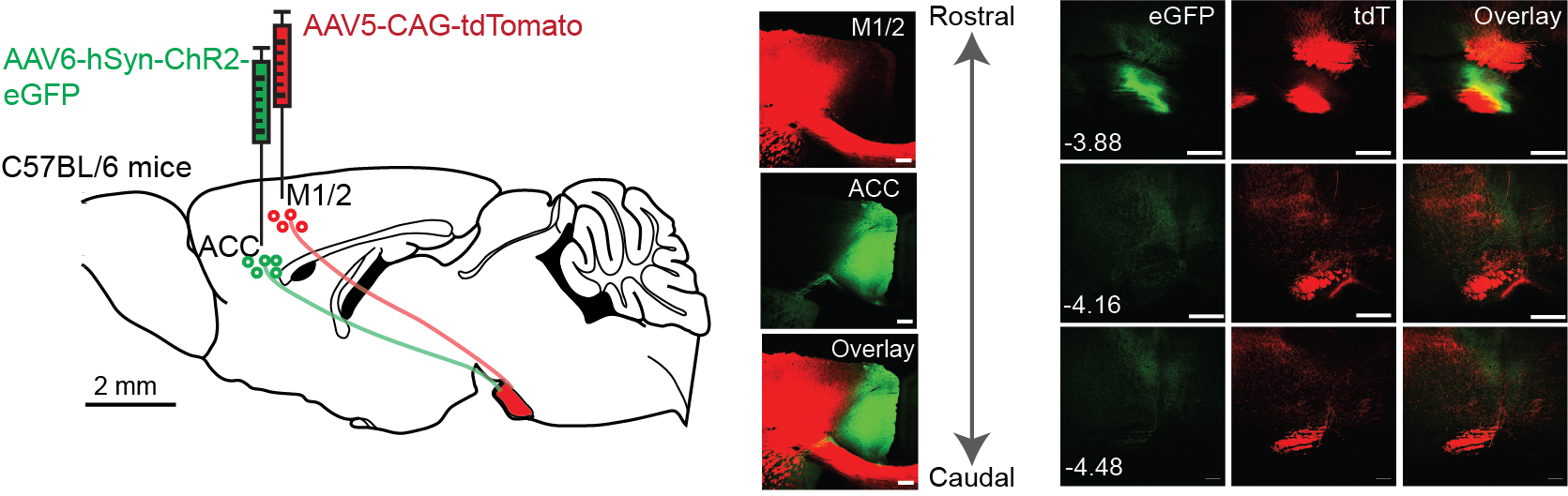
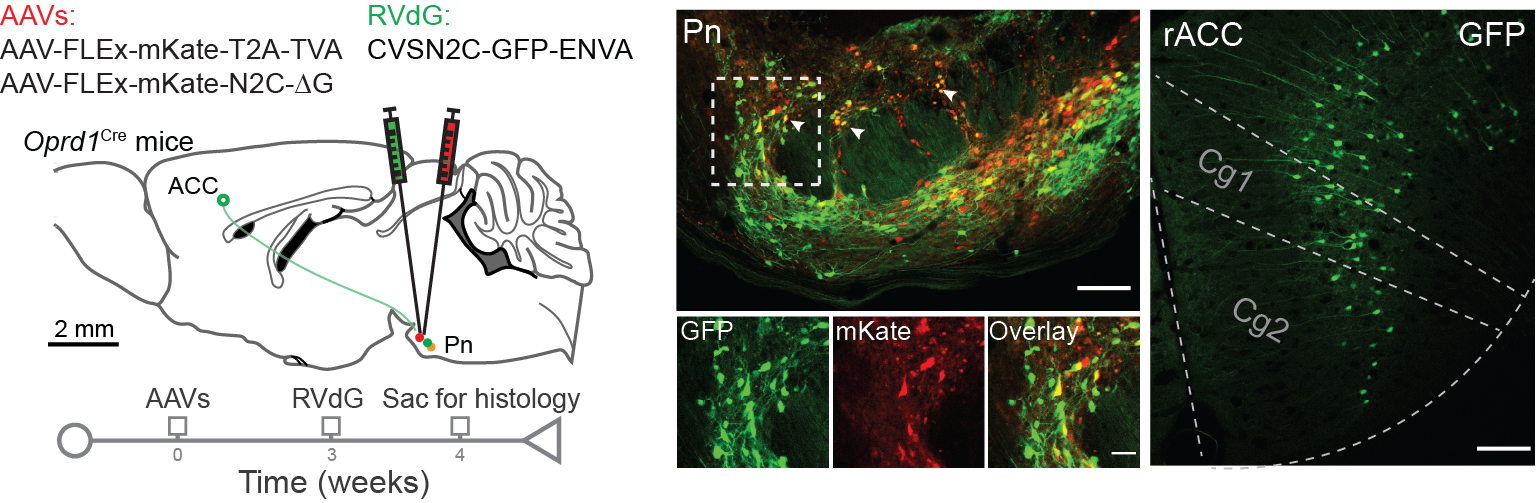
AAV-mediated anterograde tracing (left) and rabies virus-mediated retrograde tracing (right) (Chen et al., 2024).
Activity-dependent genetic labeling
Activity-dependent tagging methods, such as TRAP (Targeted Recombination in Active Populations), are cutting-edge tools for labeling and studying neurons activated during specific experiences or stimuli. TRAP uses a combination of an immediate early gene promoter, such as Fos, and CreER technology to drive conditional gene expression in active neurons upon exposure to tamoxifen. This allows for precise temporal control of labeling, capturing neurons based on their activity at specific moments. TRAP is invaluable for dissecting functional circuits, enabling researchers to manipulate, trace, or analyze activated neuronal populations, shedding light on the neural basis of behaviors, memory, and sensory processing.
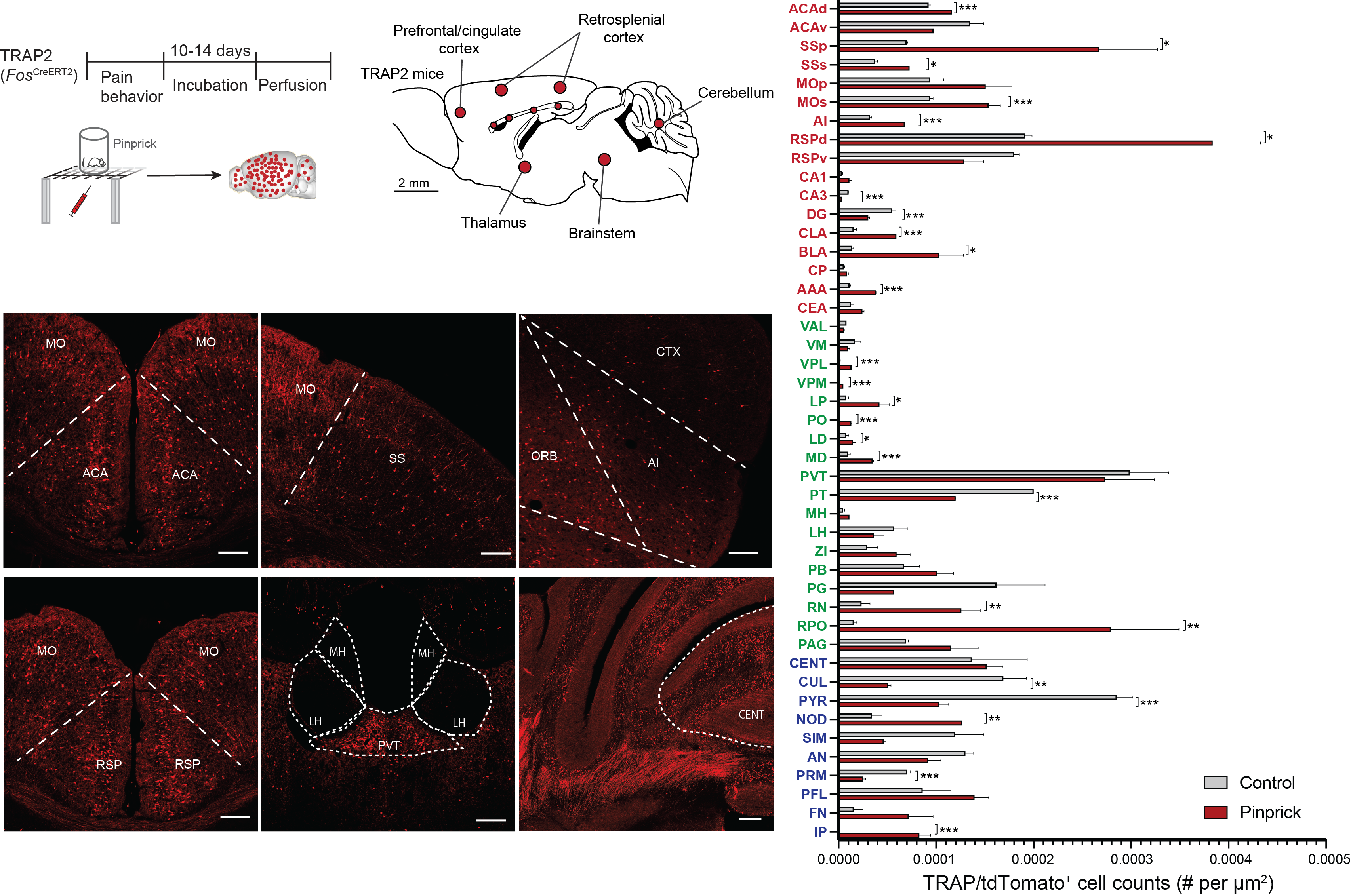
Labeling neurons active during pain stimulation using TRAP2 mice (Chen et al., 2023).
Single-cell RNA seq and RNAscope
Single-cell RNA sequencing (scRNA-seq) is a cutting-edge method for analyzing gene expression in individual cells, providing detailed insights into cellular diversity, gene networks, and functional states. By capturing and sequencing RNA from single cells, it enables the discovery of novel cell types and understanding of dynamic biological processes. In situ hybridization (ISH) is a complementary technique used to validate scRNA-seq findings by visualizing specific RNA transcripts in their spatial context within tissues. ISH provides direct evidence of gene expression at the cellular level, linking molecular data to anatomical structures and ensuring the accuracy of transcriptomic analyses.
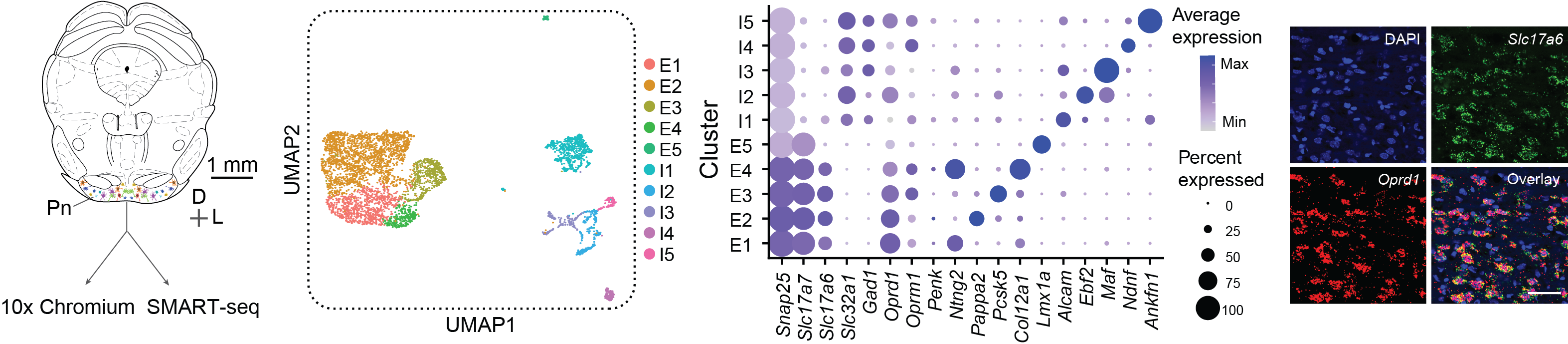
Classifying Pn neurons based on their transcriptomic profiles (Chen et al., 2024).
Optogenetics and chemogenetics
Optogenetics and chemogenetics are advanced tools for manipulating neuronal activity with high precision. Optogenetics uses light-sensitive proteins, such as channelrhodopsins and halorhodopsins, to activate or inhibit neurons via light, offering millisecond-level temporal resolution. It is widely used to probe the causal relationship between neuronal activity and behavior. Chemogenetics employs engineered receptors, such as DREADDs (Designer Receptors Exclusively Activated by Designer Drugs), which are activated by specific ligands, enabling more prolonged and spatially targeted modulation of neuronal activity. Both techniques provide valuable insights into neural circuits and behavior, with optogenetics excelling in temporal precision and chemogenetics in long-lasting effects.
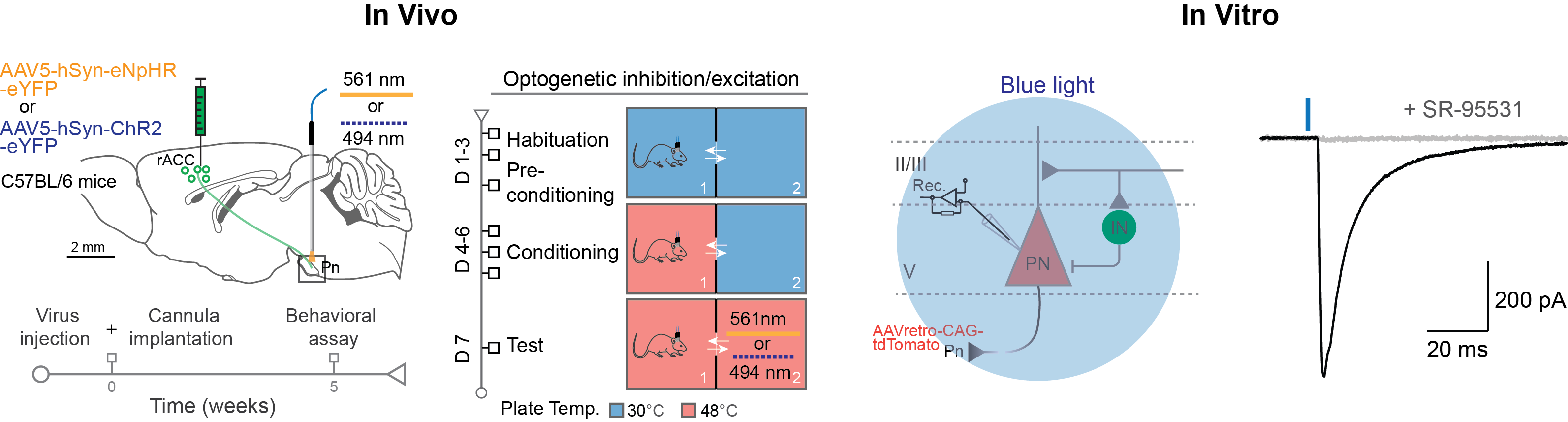
Manipulating neuronal activity using optogenetics in both in vivo and in vitro settings (Chen et al., 2024).
Computational approaches
Markerless animal pose estimation
Markerless animal pose estimation, like DeepLabCut, uses deep learning to track and analyze animal movements without requiring physical markers. It relies on annotated video data to train neural networks that predict the positions of key body parts across frames. This approach enables precise, automated tracking of behavior in diverse species and experimental setups, making it ideal for neuroscience, ethology, and biomechanics research.
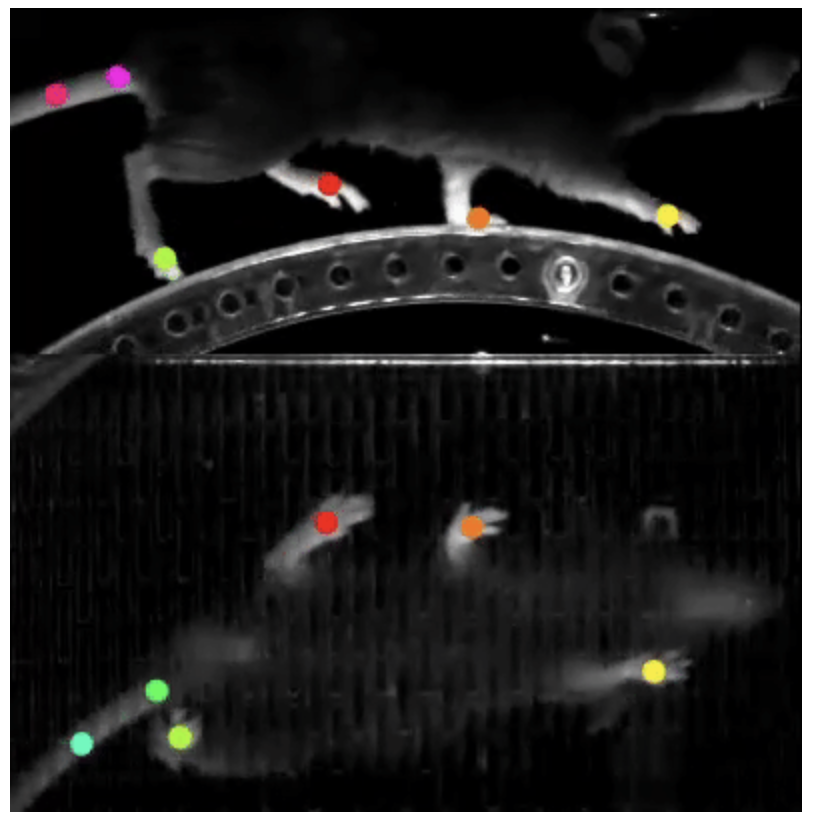
Image source: DeepLabCut official website.
Graph theory
Graph theory is a branch of mathematics focused on studying graphs, which consist of nodes (vertices) connected by edges (links). It provides tools to analyze structures and relationships in networks, such as social networks, neural connections, or transportation systems. Key concepts include connectivity, shortest paths, and centrality. In neuroscience, graph theory is used to model brain connectivity, uncovering how neural networks process information. Its versatility makes it applicable to diverse fields, from biology to computer science and beyond.
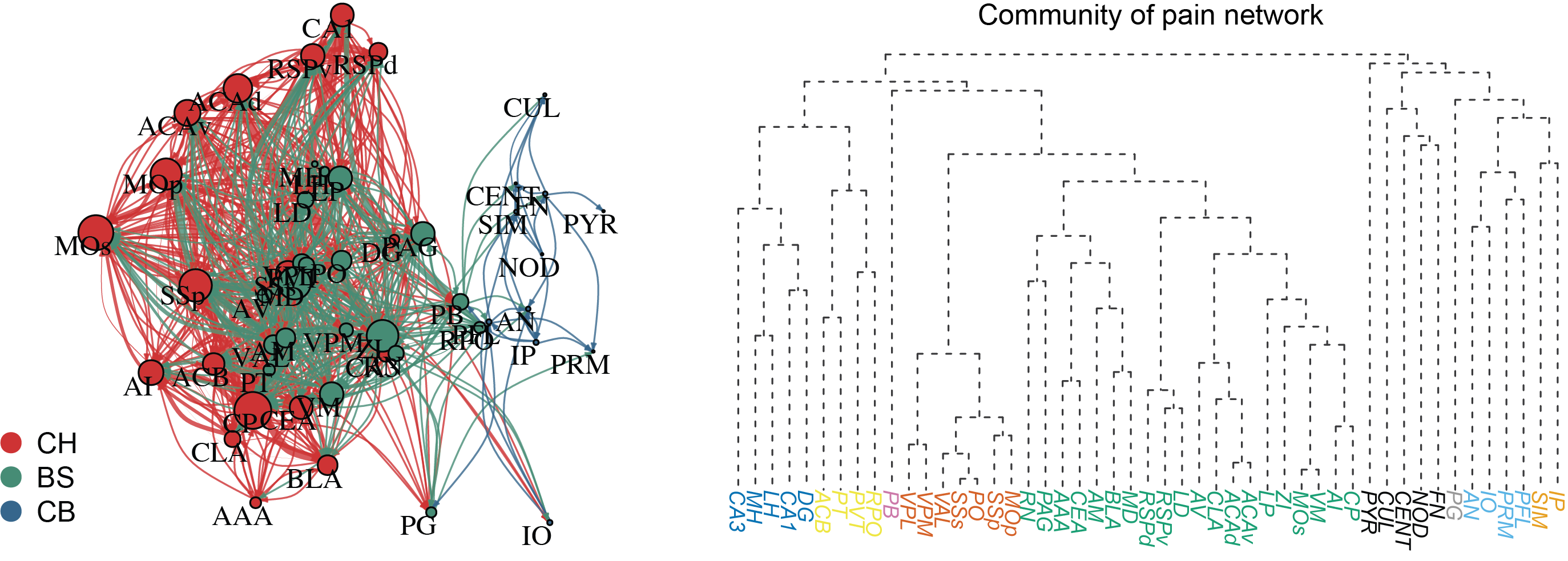
Employing graph theory to systematically characterize the architecture of a comprehensive pain network (Chen et al., 2023).
Machine learning
Machine learning (ML) and deep learning (DL) are transformative tools in neuroscience, enabling the analysis of complex datasets such as neural recordings, imaging, and behavioral data. ML algorithms, including support vector machines and random forests, are used to classify patterns and predict outcomes based on features extracted from data. Deep learning, a subset of ML, leverages artificial neural networks with multiple layers to automatically extract features and model intricate relationships. Applications in neuroscience include decoding brain activity, understanding neural connectivity, and predicting behaviors. DL excels in handling large-scale, high-dimensional data, offering unparalleled precision in unraveling the complexities of the brain.
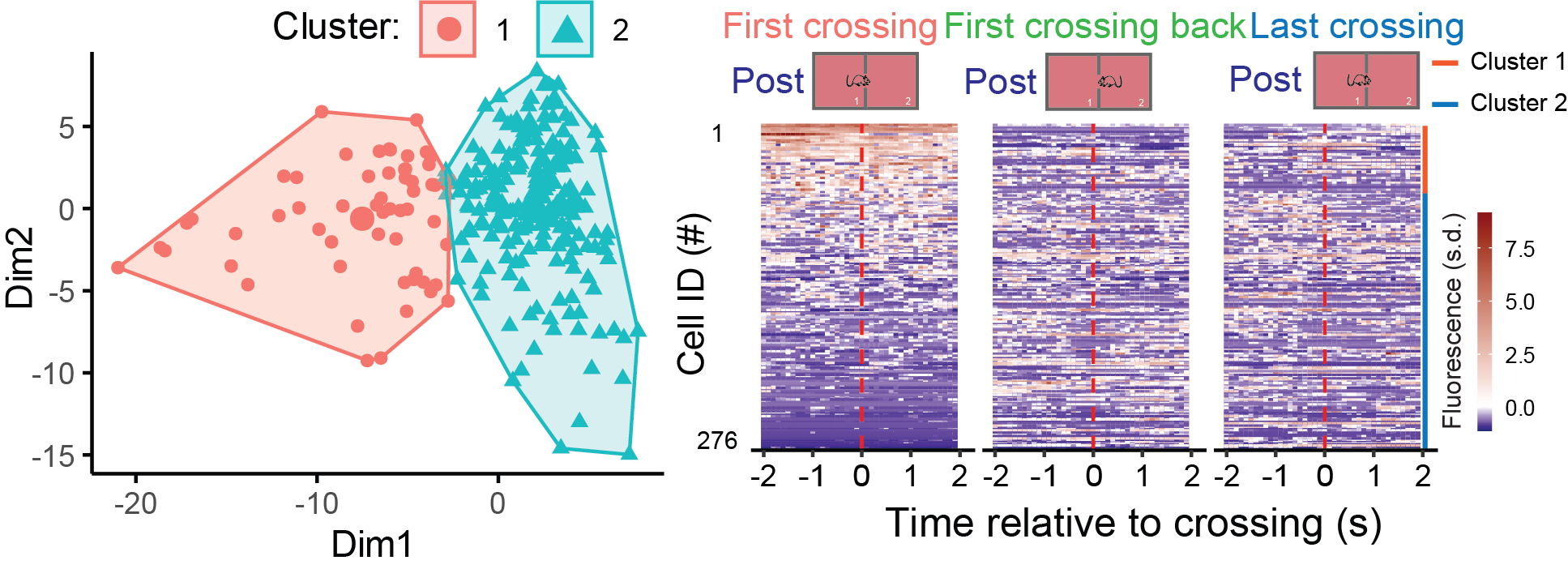
Classifying neurons based on their activity using machine learning algorithms (Chen et al., 2024).